In the past few years, remote sensing data have increasingly been used in monitoring, spatial predicting modeling, and risk assessment with respect to human health for their ability to facilitate the identification of social and environment determinants of health and to provide consistent spatial and temporal coverage of study data. In the meantime, machine learning techniques have been widely used in public health research to enhance the accuracy of disease prediction and early detection, improve the efficiency of healthcare resource allocation, and enable the development of personalized treatment plans.
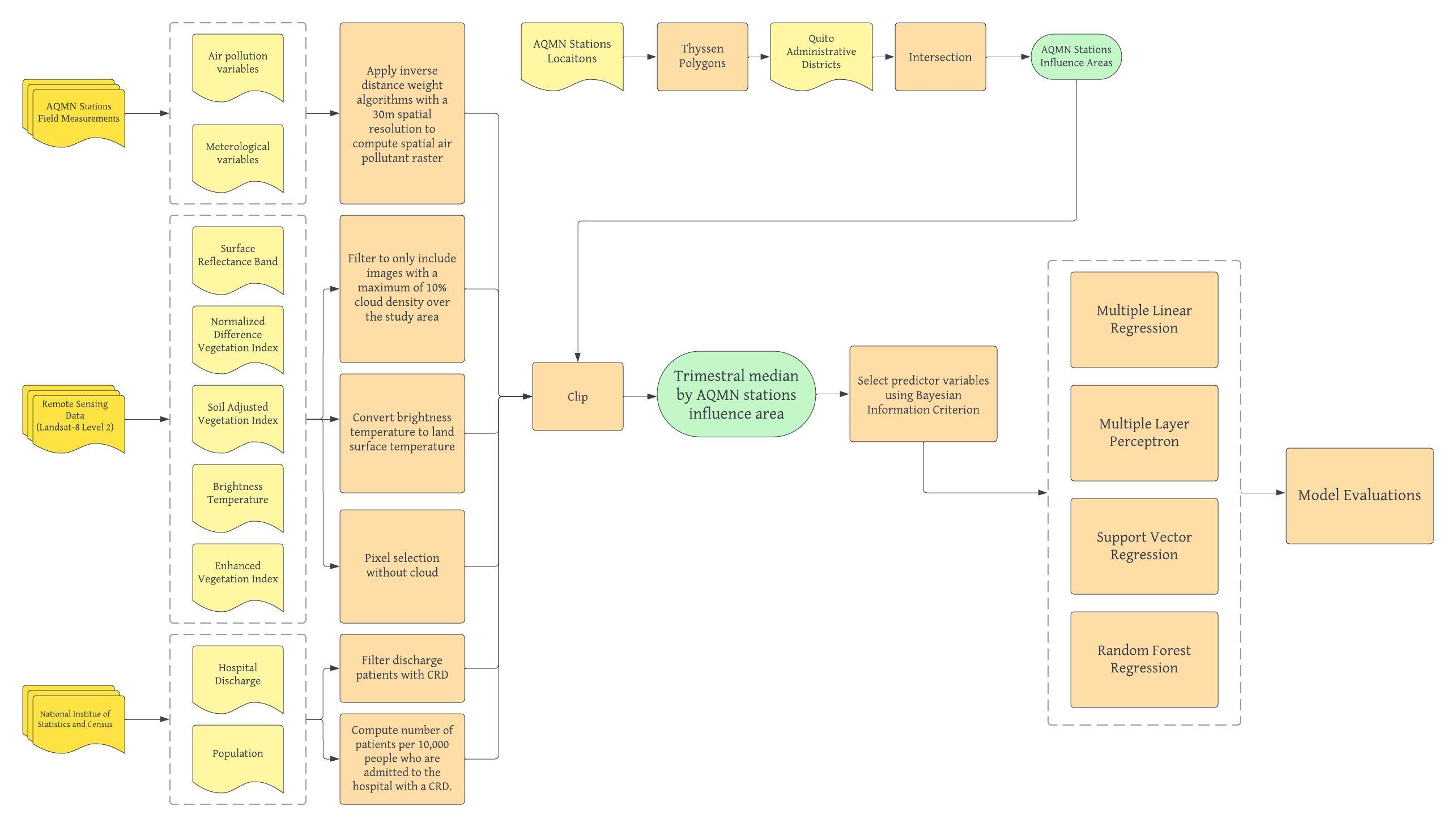
As an example of applying remote sensing data and machine learning techniques in health research, Alvarez-Mendoza et al. conducted a study in 2020, in which they proposed to estimate the prevalence of Chronic Respiratory Diseases (CRDs) by examining the relationship between remote sensing data, air quality variables, and the number of hospital discharges of patients with CRDs in Quito, Ecuador. Specifically, their study estimated and compared three different complex machine learning techniques, support vector regression, random forest regression, and multiple layer perceptron, in predicting CRD rate, considering the effect of environmental variables, air pollution field measurements, and meteorological data. Their goal is to provide insights into and an understanding of the most significant spatial predictors and the spatial distribution of HCRD in the city of Quito.
For a detailed description of Alvarez-Mendoza et al's methodology and workflow, please refer to the workflow below and the presentation slides here.
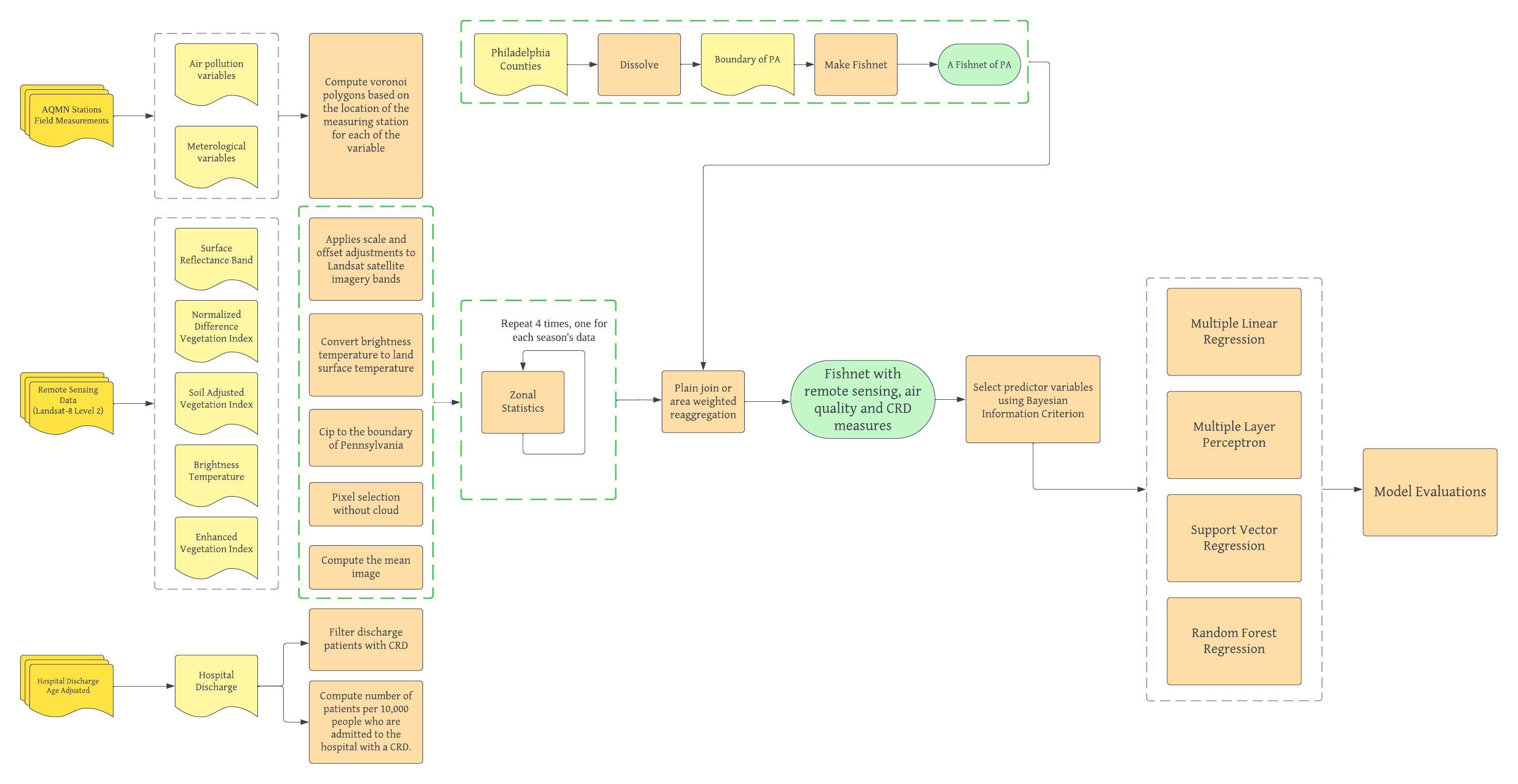
In our final project, we plan to replicate and improve upon Alvarez-Mendoza et al.’s study and investigate the effectiveness of several machine learning models in predicting the number of hospital discharge patients with CRD in the state of Pennsylvania. Following their established workflow, we combined multiple data sources, including specific bands of satellite imageries, different kinds of vegetation indices, monthly air pollutant measures, and meterological indicators, as proxies for local environment to analyze the distribution of CRD hospitalizations across the Pennsylvania. Our goal is to understand the most significant environmental and atmospheric factors leading to higher CRD risk in Pennsylvania as well as to compare the performance of different machine learning models.
Our biggest deviation from Alvarez-Mendoza et al.’s study is to conduct the analysis at a much larger geographic extent. Since our raw data all comes from different sources and with different geographic unit (eg. hospital discharge data are collected a county level while remote sensing data are at 30*30m spatial resolution), we divide Pennsylvania into 8397 5000*5000m contiguous fishnet grids. Doing so provide a regular and systematic way to divide geographic areas into smaller units. It simplifies our analysis by providing a structured framework for organizing and processing spatial data. Another major deviation from the original study is the availability of air quality data in Pennsylvania. Because the influence of air quality monitoring stations is not limited by administrative boudaries, making spatial interpolation of their influence zone becomes an important step before analysis. We computed several voronoi polygons based on station locations that collect different kinds of air quality data. Each station is associated with a polygon representing the area where it has the closet proximity, from which we could then aggregate to the fishnet. The third deviation is that we took seasonality into account while running the machine learning model. Specifically, models were ran on data from different seasons separately for cross-comparison. For reference, here is our modified workflow, with the modified part highlited in green.
This notebook documents our entire replication study and is organized into the following sections following the workflow of Alvarez-Mendoza et al.:
- Study Area: where we document the procedure of generating the fishnet and defining our area of interest.
- Remote Sensing Data: where we document our process of retreving, fine-tuning, and manipulating satellite images into format ready for further analysis. We also explains the method to calculate several indices that are used in our model.
- Hospital Discharge Data: where we document our process of cleaning hospital discharge CRD data and aggregating it to the fishnet.
- Air Quality Data: where we document our process of building voronoi polygon and around stations and aggregating station information into the fishnet.
- Dimensionality Reduction: where we ran the bayesian information criteria to select the most influential predictors of CRD in order to prevent multicolinearity and overfitting.
- Machine Learning: where we ran four machine learning models for each season and compare their accuracy.